Current Research
Section IV. Parasite differentiation and its use to study infection dynamics(II)
Once parasites differentiate form bloodstream to procyclic forms, mechanisms must operate to maintain the differentiated state. However, although there is extensive evidence for stage-regulated mRNA control, the signals responsible for this have remained cryptic. We have recently focussed on the control of molecules specifically expressed in stumpy forms. These are interesting because most genes are down-regulated or held translationally silent in stumpy forms. Hence, those that are actively translated at this stage are particularly interesting because their regulation is different from most other molecules despite being in the same physiological environment. We believe the signals that govern the expression of such genes will be informative in understanding development, rather than environmental, regulation (distinguishing them from molecules differentially expressed between bloodstream forms and tsetse midgut forms). This will help us to understand what is important for controlling transmission and hence spread of the parasite.
We have dissected the regulation of components of the differentiation regulator, PAD1, and also a novel family of molecules strongly expressed in stumpy forms, but of unknown function. These are known as expression site associated gene 9 (ESAG9). Detailed dissection of these genes has revealed that they exhibit distinct regulatory mechanisms and signals, despite their co-regulated expression.
In a new theme, pursued together with Nick Savill within our Centre for Immunity, Infection and Evolution, we have exploited the differential expression of these genes, specifically PAD1, to mathematically model the contribution of stumpy forms to the overall infection dynamics of trypanosomes in their mammalian host. This has emphasised the importance of stumpy forms in regulating the parasite load within a chronic infection and revealed how this regulates the frequency of antigenic variation in the population, thereby assisting infection longevity.
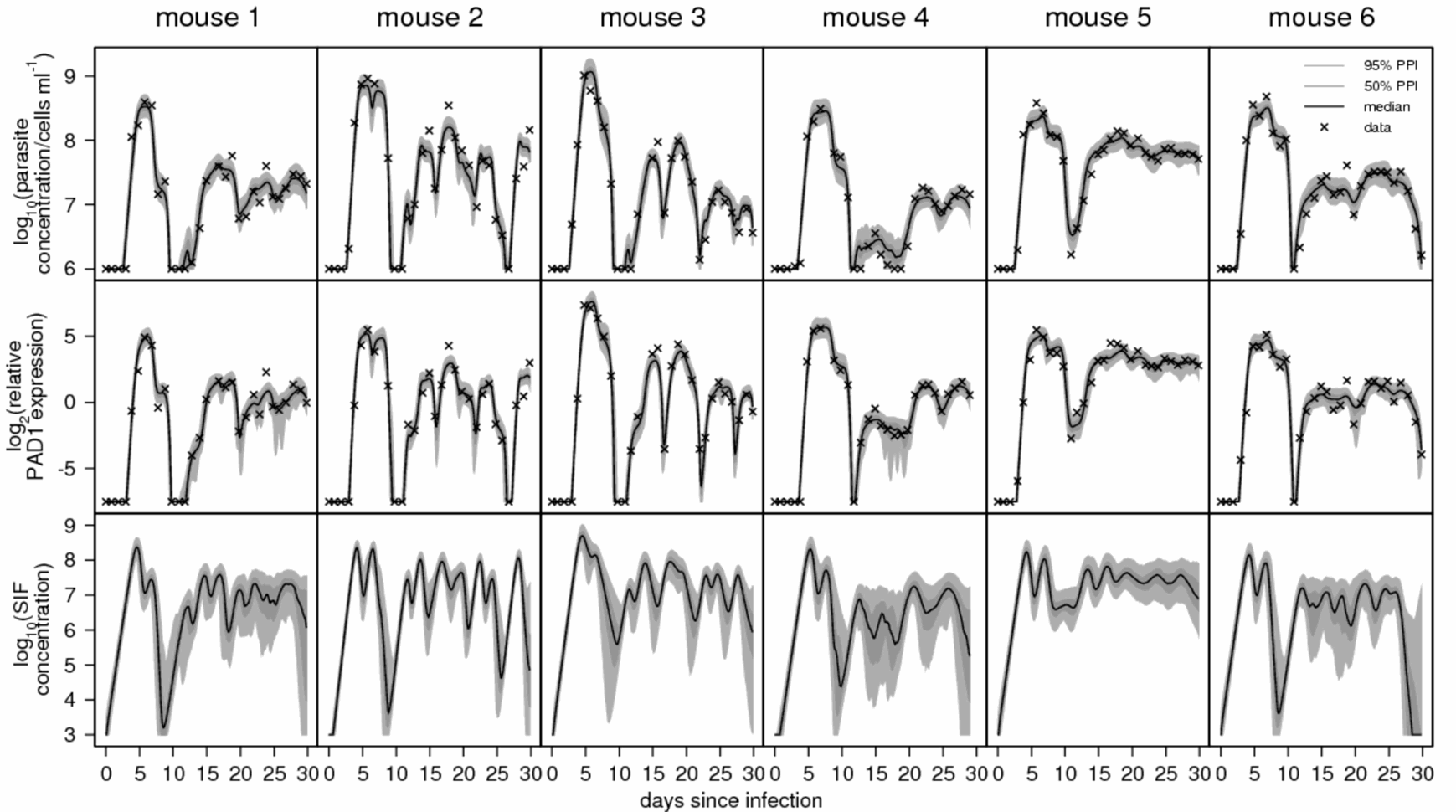
Finally, the reporter lines that accurately reflect the level of stumpy forms in the trypanosome population are being used to identify chemicals that can prematurely activate cell differentiation in the mammalian blood. We believe that this approach will help us to identify developmental accelerators, able to prevent parasite virulence. They will also assist us by providing tools as we dissect the parasites developmental signalling pathways.
For more information on this theme, see:
- Kabani S, Fenn K, Ross A, Ivens A, Smith TK, Ghazal P, Matthews K. (2009)
Genome-wide expression profiling of in vivo-derived bloodstream parasite stages and dynamic analysis of mRNA alterations during synchronous differentiation in Trypanosoma brucei. BMC Genomics. 2009 Sep 11;10:427.- MacGregor P, Matthews KR. (2010)
- New discoveries in the transmission biology of sleeping sickness parasites: applying the basics.
J Mol Med. 2010 Sep;88(9):865-71. Epub 2010 Jun 5.
Barnwell EM, van Deursen FJ, Jeacock L, Smith KA, Maizels RM, Acosta-Serrano A, Matthews K.
Developmental regulation and extracellular release of a VSG expression-site-associated gene product from Trypanosoma brucei bloodstream forms.
J Cell Sci. 2010 Oct 1;123(Pt 19):3401-11. Epub 2010 Sep 7.
MacGregor P, Savill NJ, Hall D, Matthews KR.
Transmission stages dominate trypanosome within-host dynamics during chronic infections.
Cell Host Microbe. 2011 Apr 21;9(4):310-8.
Controlling and co-ordinating development in vector-transmitted parasites
Science. Vol. 331 no. 6021 pp. 1149-1153

